A Needle in a Stack of Needles
Cells within the body are constantly being recycled. Cellular DNA is released into the peripheral bloodstream, becoming cell-free DNA (cfDNA). Non-invasive prenatal testing (NIPT) takes advantage of this fact to analyze fetal cfDNA in the mother's bloodstream for signs of aneuploidy.

Cancer tumors also shed DNA into the bloodstream. Unlike healthy cells, this DNA carries signs of mutations. Some of these mutations are responsible for the malignancy. The circulating tumor DNA (ctDNA) may be less than 0.1% of all the cfDNA. Therefore, there may be tremendous amounts of background "noise" obfuscating each meaningful signal. The fragments are all made of the same material: DNA. Therefore, it is harder than a needle in a haystack, you are actually looking for one slightly different needle in a large pile of needles.
The haploid human genome contains 3 billion bases. Some driving mutations are single nucleotide variations (SNVs), where a single base change has occurred. For example, an adenine (A) becomes a guanine (G). Some of the SNVs are heterozygous mutations, so instead of 1 in 3 billion, it's actually a mutation of 1 in 6 billions bases. If trying to reach a detection limit of 0.1% of ctDNA in 99.9% background cfDNA, the problem becomes the ability to detect one molecule in 6 trillion bases.
Complicating the analysis, the cfDNA is broken into millions of short fragments. The average length of cfDNA is only 165 bases. Purified cfDNA from plasma appears predominantly as monomeric, dimeric, and occasionally trimeric nucleosome-sized DNA fragments. These short fragments must be mapped against the human genome to find out what gene they belong to and if they contain mutations of clinical significance.
Process
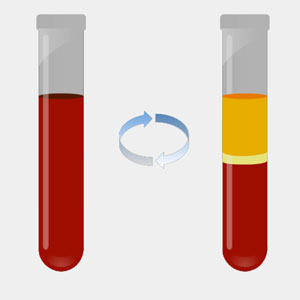
We start with a standard blood draw — a liquid biopsy. The whole blood is spun down in a centrifuge to separate plasma from the buffy coat and red-blood cell fractions. We want to eliminate leukocytes (white blood cells) as they contain DNA that if allowed to lyse, contribute greatly to the background noise. Variable amounts of high molecular weight genomic DNA found in the plasma, presumably derived from nucleated blood cells that lyse, are also observed and excluded.
The cfDNA is extracted from the plasma for analysis. Through a series of proprietary biochemical steps, we amplify the cfDNA and using very small probes, target and capture strands of ctDNA. Our novel chemistry allows for a very even and deep capture of DNA sequences, regardless of GC content.  This ability has two benefits: we can easily capture any area of the genome equally and we do not have to sequence deeply to gather enough information to make a variant call. Our ontarget rate is incredibly high, yielding > 95% sensitivity and 100% specificity. The lower level of detection is a mere 0.1% We lower the background and boost the signal to achieve exquisite sensitivity and specificity.
This ability has two benefits: we can easily capture any area of the genome equally and we do not have to sequence deeply to gather enough information to make a variant call. Our ontarget rate is incredibly high, yielding > 95% sensitivity and 100% specificity. The lower level of detection is a mere 0.1% We lower the background and boost the signal to achieve exquisite sensitivity and specificity.
Unlike PCR-based methods, our system can discover mutations. We do not need to "pre-program" specific variant sequences prior to capture. The Resolution platform can find SNPs, insertion and deletions, copy number variations (CNVs) / amplifications, and fusions / translocations. Critically, we can find fusions without any prior knowledge of the fusion partner or specific breakpoint.
The extreme efficiency of the Resolution Bio ctDx platform allows us to analyze multiple patient samples on a single desktop sequencer run. The resultant data is just a few GBs — up to 250x less than some other methods. The result is faster turn around time and lower cost.
We currently run assays within our CLIA lab. We are working hard to create kits of our biochemistry, so labs around the world will be able to quickly and efficiently return clinically actionable results to clinicians.
If you would like more information, please contact us via the inquiry form.
Proprietary biochemistry molecularly annotates every cfDNA fragment. The chemistry informs our custom, cloud-based bioinformatics pipeline which generates the variant report.