Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients
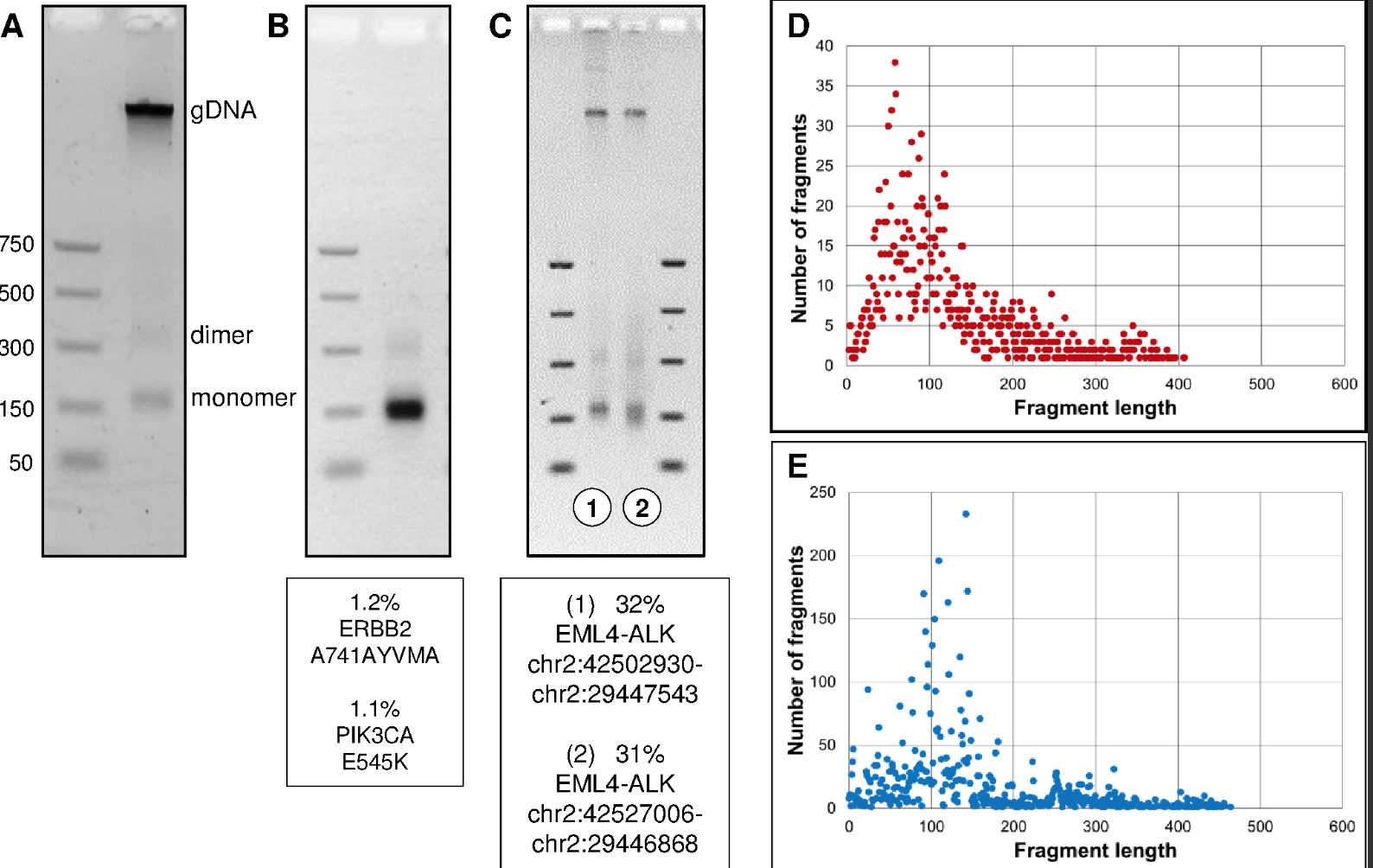
Purpose: Tumor genotyping is a powerful tool for guiding non-small cell lung cancer (NSCLC) care, however comprehensive tumor genotyping can be logistically cumbersome. To facilitate genotyping, we developed a next-generation sequencing (NGS) assay using a desktop sequencer to detect actionable mutations and rearrangements in cell-free plasma DNA (cfDNA). Experimental Design: An NGS panel was developed targeting 11 NSCLC driver oncogenes. Targeted NGS was performed using a novel methodology that maximizes on-target reads, and minimizes artifact, and validated on DNA dilutions derived from cell lines. Plasma NGS was then blindly performed on 48 patients with advanced, progressive NSCLC and a known genotype, and explored in two patients with incomplete tumor genotyping. Results: NGS could identify mutations present in DNA dilutions at ≥0.4% allelic frequency with 100% sensitivity/specificity. Plasma NGS detected a broad range of driver and resistance mutations, including ALK rearrangements, HER2 insertions, and MET amplification, with 100% specificity. Sensitivity was 77% across 62 known driver and resistance mutations from the 48 cases; in 29 cases with common EGFR and KRAS mutations, sensitivity was similar to droplet digital PCR. In two cases with incomplete tumor genotyping, plasma NGS rapidly identified a novel EGFR exon 19 deletion and a missed case of MET amplification. Conclusion: Blinded to tumor genotype, this plasma NGS approach detected targetable genomic alterations in NSCLC with no false positives. Through use of widely available vacutainers and a desktop sequencing platform, this assay has the potential to be implemented broadly for patient care and translational research.
Read More